The continuation of the research has focused
on the two most significantly inhibited microbes, the E.coli (EC) and the
L.monocytogenes (LM). For the other three, last week’s experiment is repeated
on them, using fresh culture.
2.1 For EC and LM, three concentrations are
prepared; 100%, 50% and 25%. The original sample is diluted until the desired
concentration.
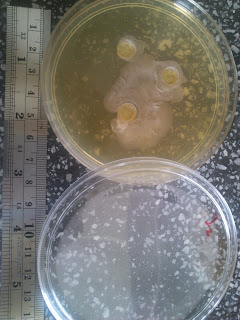
Result is as follows:
a) LM
Conc.
|
50
|
25
|
1
|
12
|
15
|
2
|
12
|
0
|
3
|
13
|
0
|
AVERAGE
|
12.3
|
15
|
b) EC
Conc.
|
50
|
25
|
1
|
0
|
12.5
|
2
|
0
|
12
|
3
|
0
|
12
|
AVERAGE
|
0
|
12.16667
|
Main problem: Sample is too viscous and sticky
to be removed using a pipette. Therefore solvent is poured in and shaken as
much as possible to dissolve the sample. But not all is dissolved, some remain
sticking to the base of the container.
Therefore it is difficult to gauge the real
inhibitory activity for 25% and 50% since there might actually be lower concentration
of ciku compound in diluted sample.
2.2 For Retest of antimicrobial susceptibility
(SA, STY and I), result is as follows:
Take note that if a “ring” is observed for streptococci
types, it is deemed resistant regardless of the clear zone size, because of
the beta-lactamase produced by the m/o.
Suggestion:
to do it in a spread plate method, using cotton swabs, to make sure the m/o spreads properly.
Also, look into other journals, to see what kind of extract is used--eg. methanol and hexane, since the viscosity may be attributed to the solvent.
Suggestion:
to do it in a spread plate method, using cotton swabs, to make sure the m/o spreads properly.
Also, look into other journals, to see what kind of extract is used--eg. methanol and hexane, since the viscosity may be attributed to the solvent.